-
Study
-
Quick Links
- Open Days & Events
- Real-World Learning
- Unlock Your Potential
- Tuition Fees, Funding & Scholarships
- Real World Learning
-
Undergraduate
- Application Guides
- UCAS Exhibitions
- Extended Degrees
- School & College Outreach
- Information for Parents
-
Postgraduate
- Application Guide
- Postgraduate Research Degrees
- Flexible Learning
- Change Direction
- Register your Interest
-
Student Life
- Students' Union
- The Hub - Student Blog
- Accommodation
- Northumbria Sport
- Support for Students
-
Learning Experience
- Real-World Learning
- Research-enriched learning
- Graduate Futures
- The Business Clinic
- Study Abroad
-
-
International
International
Northumbria’s global footprint touches every continent across the world, through our global partnerships across 17 institutions in 10 countries, to our 277,000 strong alumni community and 150 recruitment partners – we prepare our students for the challenges of tomorrow. Discover more about how to join Northumbria’s global family or our partnerships.
View our Global Footprint-
Quick Links
- Course Search
- Undergraduate Study
- Postgraduate Study
- Information for Parents
- London Campus
- Northumbria Pathway
- Cost of Living
- Sign up for Information
-
International Students
- Information for International Students
- Northumbria and your Country
- International Events
- Application Guide
- Entry Requirements and Education Country Agents
- Global Offices and Regional Teams
- English Requirements
- English Language Centre
- International student support
- Cost of Living
-
International Fees and Funding
- International Undergraduate Fees
- International Undergraduate Funding
- International Masters Fees
- International Masters Funding
- International Postgraduate Research Fees
- International Postgraduate Research Funding
- Useful Financial Information
-
International Partners
- Agent and Representatives Network
- Global Partnerships
- Global Community
-
International Mobility
- Study Abroad
- Information for Incoming Exchange Students
-
-
Business
Business
The world is changing faster than ever before. The future is there to be won by organisations who find ways to turn today's possibilities into tomorrows competitive edge. In a connected world, collaboration can be the key to success.
More on our Business Services-
Business Quick Links
- Contact Us
- Business Events
- Research and Consultancy
- Education and Training
- Workforce Development Courses
- Join our mailing list
-
Education and Training
- Higher and Degree Apprenticeships
- Continuing Professional Development
- Apprenticeship Fees & Funding
- Apprenticeship FAQs
- How to Develop an Apprentice
- Apprenticeship Vacancies
- Enquire Now
-
Research and Consultancy
- Space
- Energy
- AI Futures
- CHASE: Centre for Health and Social Equity
- NESST
-
-
Research
Research
Northumbria is a research-rich, business-focused, professional university with a global reputation for academic quality. We conduct ground-breaking research that is responsive to the science & technology, health & well being, economic and social and arts & cultural needs for the communities
Discover more about our Research-
Quick Links
- Research Peaks of Excellence
- Academic Departments
- Research Staff
- Postgraduate Research Studentships
- Research Events
-
Research at Northumbria
- Interdisciplinary Research Themes
- Research Impact
- REF
- Partners and Collaborators
-
Support for Researchers
- Research and Innovation Services Staff
- Researcher Development and Training
- Ethics, Integrity, and Trusted Research
- University Library
- Vice Chancellors Fellows
-
Research Degrees
- Postgraduate Research Overview
- Doctoral Training Partnerships and Centres
- Academic Departments
-
Research Culture
- Research Culture
- Research Culture Action Plan
- Concordats and Commitments
-
-
About Us
-
About Northumbria
- Our Strategy
- Our Staff
- Our Schools
- Place and Partnerships
- Leadership & Governance
- University Services
- Northumbria History
- Contact us
- Online Shop
-
-
Alumni
Alumni
Northumbria University is renowned for the calibre of its business-ready graduates. Our alumni network has over 253,000 graduates based in 178 countries worldwide in a range of sectors, our alumni are making a real impact on the world.
Our Alumni - Work For Us
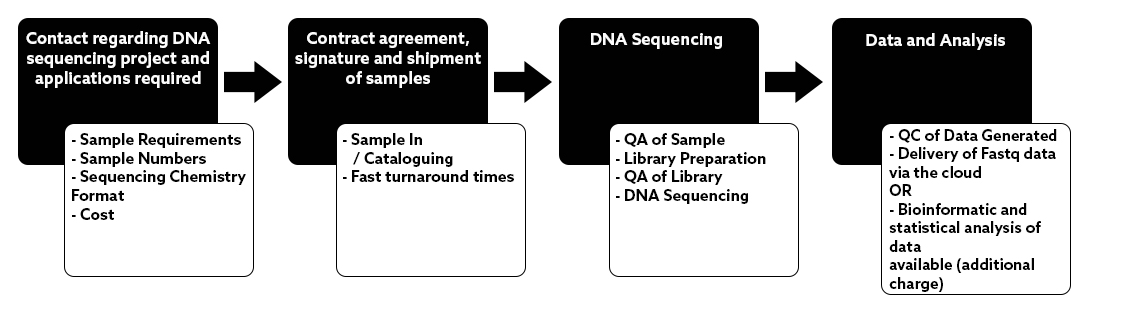
 NU-OMICS was one of the first sites in the UK to install the Illumina Miseq in 2013.
NU-OMICS was one of the first sites in the UK to install the Illumina Miseq in 2013.
We have since built significant experience in small genome sequencing and microbiome studies using Illumina sequencing by synthesis.
Sample preparations are automated using our Hamilton Star liquid handling system. We are not restricted by target for amplicon sequencing. We can help validate new amplicon targets for microbial community sequencing and our academic led facility can support novel approaches or new technologies.
Please ask if you have a project that is not listed in the application section.
Bacterial community amplicon sequencing
- Multiple Amplicon targets
- 16S rRNA gene sequencing V4 (multiple versions).
- We have validated studies with other primer targets. Please ask if your target-of-interest could be used for bacterial community projects.
Fungal community amplicon sequencing
- Multiple Amplicon targets
- ITS1-2
- We have validated studies with other primer targets, so please ask if your target-of-interest could be used for fungal community projects.
Small genome sequencing
- Bacteria
- Nextera XT
- Eukaryotic viruses
- Nextera XT
- Bacteriophages
- Nextera XT
Shotgun Metagenomics
- Untargeted
- Viral metagenomics DNA and RNA viruses.
- Methods and bioinformatics designed by academic team.
Bacterial transcriptomics
- Using ScritpSeq Library Preparation sequencing kit
TruSeq, panel/design for eukaryotic gene expression
Contact for further details or follow the below link to find validated Illumina panels.
https://emea.illumina.com/products/selection-tools/gene-panel-finder.html#/targeted-panels
miRNA sequencing
- TruSeq Small RNA sequencing
Please send samples to the following delivery address;
NU-OMICS DNA sequencing research facility
(C/O) Darren Smith/Andrew Nelson
Northumbria University
Ellison Building EBA 504a
Newcastle Upon Tyne
Tyne and Wear
NE1 8ST
Small Genome sequencing; Bacterial or virus
- NexteraXT
- All samples must be treated with broad spectrum RNAse prior to sending. An electrophoresis image is required for QA purposes run on a 1% agarose gel. Limited shearing should be present, if worried contact us to check before sending.
- All samples should be in a 96 well format as parts of the preparation are automated. Data will be returned labelled in well-format over sample name. DNA quality A260/280 should be >1.8.
- For Nextera XT we require a minimum of 55 ng of DNA in 11ul of elution buffer.
Bacterial and Fungal Community amplicon sequencing
- DNA sample extraction should be with either phenol:chloroform or for kit based methods we recommend the Qiagen (MoBio) PowerSoil DNA extraction kit.
- All samples should be in a 96 well format as parts of the preparation are automated. Data will be returned labelled in well-format over sample name. DNA quality A260/280 should be >1.8. Please trial the DNA extraction to see if possible to amplify the target gene in a subset of your samples.
- Always include a DNA extraction negative. If you use an extraction kit or require multiple kits due to sample numbers please provide a DNA extraction negative for each kit. We will provide a sequencing negative and a commercial mock community so that downstream analysis can be completed (https://www.zymoresearch.eu/zymobiomics-community-standard)
Bacterial RNA sequencing
- Using ScritpSeq Library Preparation sequencing kit
- We require a minimum of 1ug of total RNA with RIN value >1.8, although higher is recommended. We can provide QA of total RNA extraction at an extra cost.
- We are happy to accept libraries already prepared for pooling as long as the supporting QA is available.
Shotgun metagenomics
- NexteraXT
- All samples must be treated with broad spectrum RNAse prior to sending. An electrophoresis image is required for QA purposes run on a 1% agarose gel. Limited shearing should be present, if worried email Nuomics@northumbria.ac.uk to check before sending.
- All samples should be in a 96 well format as parts of the preparation are automated. Data will be returned labelled in well-format over sample name. DNA quality A260/280 should be >1.8.
- For Nextera XT we require a minimum of 55 ng of DNA in 11ul of elution buffer.
Targeted RNA sequencing from predesigned or bespoke amplicon panels (TruSeq)
- We require a minimum of 1ug of total RNA with RIN value >1.8, although higher is recommended.
- We are happy to accept libraries already prepared for pooling as long as the supporting QA is available.
Small RNA sequencing
- We require a minimum of 1ug of total RNA with RIN value >1.8, although higher is recommended.